I just want to show another two plots for the statistical power of a test, since I didn't have time for this earlier
The code to produce them is just calling the methods of the power classes, for example for the one sample t-test.
I just want to show another two plots for the statistical power of a test, since I didn't have time for this earlier
The code to produce them is just calling the methods of the power classes, for example for the one sample t-test.
I think I must be turning too much into a statistician and econometrician lately, I must have caught a virus or something. Maybe it started already a while ago
The theme of the scipy conference this year is "Machine Learning & Tools for Reproducible Science". However, I'm not doing any sexy twitter analysis, I just spent some days coding tests for proportion, boring stuff like pairwise comparisons of proportions.
Anyway, I decided to submit a tutorial proposal for econometrics with statsmodels to the scipy conference, see (lightly edited) proposal below. Since my proposal didn't get accepted, my first response was: Wrong topic, Too much statistic, We just want numbers, not check whether the model is correct, and find out how to fix it.
That leaves me with more time to go back to figuring out which other basic statistical tests are still missing in Python.
This is just a short comment on a blog post.
Ferando Perez wrote a nice article about "Literate computing" and computational reproducibility: IPython in the age of data-driven journalism
In the second part, he explains that Vincent Arel-Bundock came up with an ipython notebook within three hours to replicate some criticism of an economics journal article. Vincent's notebook can be seen here.
What I found most striking was not the presentation as a notebook, although that makes it easy to read, instead it was: pandas, patsy and statsmodels, and no R in sight. We have come a long way with Statistics in Python since I started to get involved in it five years ago.
Vincent has made many improvements and contributions to statsmodels in the last year.
Aside
I'm not following much of the economics debates these days, so I only know what I read in the two references that Fernando gave.
My impression is that it's just the usual (mis)use of economics research results. Politicians like the numbers that give them ammunition for their position. As economist, you are either very careful about how to present the results, or you join the political game (I worked for several years in an agricultural department of a small country). (An example for the use of economics results in another area, blaming the financial crisis on the work on copulas.)
"Believable" research: If your results sound too good or too interesting to be true, maybe they are not, and you better check your calculations. Although mistakes are not uncommon, the business as usual part is that the results are often very sensitive to assumptions, and it takes time to figure out what results are robust. I have seen enough economic debates where there never was a clear answer that convinced more than half of all economists. A long time ago, when the Asian Tigers where still tigers, one question was: Did they grow because of or in spite of government intervention?
series : "Statistics in Python"
This is a follow-up to my previous posts, here and this post, which are on software development, and multiple comparisons which looks at a specific case of pairwise mean comparisons.
In the following, I provide a brief introduction to the statsmodels functions for p-value asjustements to correct for multiple testing problems and then illustrate some properties using several Monte Carlo experiments.
subtitle: "The earth is round after all"
series : "Adventures with Statistics in Python"
If you run an experiment and it shows that the earth is not round, then you better check your experiment, your instrument, and don't forget to look up the definition of "round"
Statsmodels has 11 methods for correcting p-values to take account of multiple testing (or it will have after I merge my latest changes).
The following mainly describes how it took me some time to figure out what the interpretation of the results of a Monte Carlo run is. I wrote the Monte Carlo to verify that the multiple testing p-value corrections make sense. I will provide some additional explanations of the multiple testing function in statsmodels in a follow-up post.
Let's start with the earth that doesn't look round.
The first results
==========================================================================================
b s sh hs h hommel fdr_i fdr_n fdr_tsbky fdr_tsbh fdr_gbs
------------------------------------------------------------------------------------------
reject 9.6118 9.619 9.7178 9.7274 9.7178 9.72 10.3128 9.8724 10.5152 10.5474 10.5328
r_>k 0.0236 0.0246 0.0374 0.0384 0.0374 0.0376 0.2908 0.0736 0.3962 0.4118 0.4022
------------------------------------------------------------------------------------------
The headers are shortcuts for the p-value correction method. In the first line, reject, are the average number of rejections across Monte Carlo iterations. The second line, r_>k, are the fraction of cases where we reject more than 10 hypothesis. The average number of rejections is large because the alternative in the simulation is far away from the null hypothesis, and the corresponding p-values are small. So all methods are able to reject most of the false hypotheses.
The last three methods estimate, as part of the algorithm, the number of null hypotheses that are correct. All three of those methods reject a true null hypothesis in roughly 40% of all cases. All methods are supposed to limit the false discovery rate (FDR) to alpha which is 5% in this simulations. I expected the fraction in the last line to be below 0.05. So what's wrong?
It looks obvious, after the fact, but it had me puzzled for 3 days.
Changing the experiment: The above data are based on p-values that are the outcome of 30 independent t-tests, which is already my second version for generating random p-values. For my third version, I changed to a data generating process similar to Benjamini, Krieger and Yekutieli 2006, which is the article on which fdr_tsbky is based. None of the changes makes a qualitative difference to the results.
Checking the instrument: All p-values corrections except fdr_tsbky and fdr_gbs are tested against R. For the case at hand, the p-values for fdr_tsbh are tested against R's multtest package. However, the first step is a discrete estimate (number of true null hypothesis) and since it is discrete, the tests will not find differences that show up only in borderline cases. I checked a few more cases which also verify against R. Also, most methods have a double implementation, separately for the p-value correction and for the rejection boolean. Since they all give identical or similar answers, I start to doubt that there is a problem with the instrument.
Is the earth really round? I try to read through the proof that these adaptive methods limit the FDR to alpha, to see if I missed some assumptions, but give up quickly. These are famous authors, and papers that have long been accepted and been widely used. I also don't find any assumption besides independence of the p-values, which I have in my Monte Carlo. However, looking a bit more closely at the proofs shows that I don't really understand FDR. When I implemented these functions, I focused on the algorithms and only skimmed the interpretation.
What is the False Discovery Rate? Got it. I should not rely on vague memories of definitions that I read two years ago. What I was looking at, is not the FDR.
One of my new results (with a different data generating process in the Monte Carlo, but still 10 out of 30 hypotheses are false)
==============================================================================================
b s sh hs h hommel fdr_i fdr_n fdr_tsbky fdr_tsbh fdr_gbs
----------------------------------------------------------------------------------------------
reject 5.2924 5.3264 5.5316 5.5576 5.5272 5.5818 8.1904 5.8318 8.5982 8.692 8.633
rejecta 5.2596 5.2926 5.492 5.5176 5.488 5.5408 7.876 5.7744 8.162 8.23 8.1804
reject0 0.0328 0.0338 0.0396 0.04 0.0392 0.041 0.3144 0.0574 0.4362 0.462 0.4526
r_>k 0.0002 0.0002 0.0006 0.0006 0.0006 0.0006 0.0636 0.0016 0.1224 0.1344 0.1308
fdr 0.0057 0.0058 0.0065 0.0065 0.0064 0.0067 0.0336 0.0081 0.0438 0.046 0.0451
----------------------------------------------------------------------------------------------
reject : average number of rejections
rejecta : average number of rejections for cases where null hypotheses is false (10)
rejecta : average number of rejections for cases where null hypotheses is true (20)
r_>k : fraction of Monte Carlo iterations where we reject more than 10 hypotheses
fdr : average of the fraction of rejections when null is true out of all rejections
The last numbers look much better, the numbers are below alpha=0.05 as required, including the fdr for the last three methods.
"Consider the problem of testing m null hypotheses h1, ..., hm simultaneously, of which m0 are true nulls. The proportion of true null hypotheses is denoted by mu0 = m0/m. Benjamini and Hochberg(1995) used R and V to denote, respectively, the total number of rejections and the number of false rejections, and this notation has persisted in the literature. <...> The FDR was loosely defined by Benjamini and Hochberg(1995) as E(V/R) where V/R is interpreted as zero if R = 0." Benjamini, Krieger and Yekutieli 2006, page 2127
Some additional explanations are in this Wikipedia page
What I had in mind when I wrote the code for my Monte Carlo results, was the family wise error rate, FWER,
"The FWER is the probability of making even one type I error in the family, FWER = Pr(V >= 1)" Wikipedia
Although, I did not look up that definition either. What I actually used, is Pr(R > k) where k is the number of false hypothesis in the data generating process. Although, I had chosen my initial cases so Pr(R > k) is close to Pr(V > 0).
In the follow-up post I will go over the new Monte Carlo results, which now look all pretty good.
Reference
Benjamini, Yoav, Abba M. Krieger, and Daniel Yekutieli. 2006. “Adaptive Linear Step-up Procedures That Control the False Discovery Rate.” Biometrika 93 (3) (September 1): 491–507. doi:10.1093/biomet/93.3.491.
This is part three in adventures with statsmodels.stats, after power and multicomparison.
This time it is about Cohen's Kappa, a measure of inter-rater agreement or reliability. Suppose we have two raters that each assigns the same subjects or objects to one of a fixed number of categories. The question then is: How well do the raters agree in their assignments? Kappa provides a measure of association, the largest possible value is one, the smallest is minus 1, and it has a corresponding statistical test for the hypothesis that agreement is only by chance. Cohen's kappa and similar measures have widespread use, among other fields, in medical or biostatistic. In one class of applications, raters are doctors, subjects are patients and the categories are medical diagnosis. Cohen's Kappa provides a measure how well the two sets of diagnoses agree, and a hypothesis test whether the agreement is purely random.
For more background see this Wikipedia page which was the starting point for my code.
Most of the following focuses on weighted kappa, and the interpretation of different weighting schemes. In the last part, I add some comments about R, which provided me with several hours of debugging, since I'm essentially an R newbie and have not yet figured out some of it's "funny" behavior.
Statistical tests are often grouped into one-sample, two-sample and k-sample tests, depending on how many samples are involved in the test. In k-sample tests the usual Null hypothesis is that a statistic, for example the mean as in a one-way ANOVA, or the distribution in goodness-of-fit tests, is the same in all groups or samples. The common test is the joint test that all samples have the same value, against the alternative that at least one sample or group has a different value.
However, often we are not just interested in the joint hypothesis if all samples are the same, but we would also like to know for which pairs of samples the hypothesis of equal values is rejected. In this case we conduct several tests at the same time, one test for each pair of samples.
This results, as a consequence, in a multiple testing problem and we should correct our test distribution or p-values to account for this.
I mentioned some of the one- and two sample test in statsmodels before. Today, I just want to look at pairwise comparison of means. We have k samples and we want to test for each pair whether the mean is the same, or not.
Instead of adding more explanations here, I just want to point to R tutorial and also the brief description on Wikipedia. A search for "Tukey HSD" or multiple comparison on the internet will find many tutorials and explanations.
The following are examples in statsmodels and R interspersed with a few explanatory comments.
I merged last week a branch of mine into statsmodels that contains large parts of basic power calculations and some effect size calculations. The documentation is in this section . Some parts are still missing but I thought I have worked enough on this for a while.
(Adding the power calculation for a new test now takes approximately: 3 lines of real code, 200 lines of wrapping it with mostly boiler plate and docstrings, and 30 to 100 lines of tests.)
The first part contains some information on the implementation. In the second part, I compare the calls to the function in the R pwr package to the calls in my (statsmodels') version.
I am comparing it to the pwr package because I ended up writing almost all unit tests against it. The initial development was based on the SAS manual, I used the explanations on the G-Power website for F-tests, and some parts were initially written based on articles that I read. However, during testing I adjusted the options (and fixed bugs), so I was able to match the results to pwr, and I think pwr has just the right level of abstraction and easiness of use, that I ended up with code that is pretty close to it.
If you just want to see the examples, skip to the second part.
payoff_s1 = sigma * np.random.randn(nobs) payoff_s2 = 0.1 + sigma * np.random.randn(nobs)I picked sigma=0.5 to get good numbers for the story.
stats.ttest_ind(payoff_s1, payoff_s2) smws.tost_ind(payoff_s1, payoff_s2, -0.5, 0.5, usevar='pooled')The null hypothesis for the t-test is that the two samples have the same mean. If the p-value of the t-test is below, say 0.05, we reject the hypothesis that the two means are the same. If the p-value is above 0.05, then we don't have enough evidence to reject the null hypothesis. This can also happen when the power of the test is not high enough given our sample size.
nobs: 10 diff in means: -0.14039151695 ttest: 0.606109617438 not different tost: 0.0977715582206 differentWith 10 observations the information is not enough to reject the null hypothesis in either test. The t-test says we cannot reject that they are different. The TOST test says we cannot reject that they are the same.
nobs: 100 diff in means: 0.131634043864 ttest: 0.0757146249227 not different tost: 6.39909387346e-07 not differentThe t-test does not reject that they are the same at a significance size of 0.05. The TOST test now rejects the hypothesis that there is a large (at least 0.5) difference.
nobs: 1000 diff in means: 0.107020981612 ttest: 1.51161249802e-06 different tost: 1.23092818968e-65 not differentBoth tests no reject their null hypothesis. The t-test rejects that the means are the same. However the mean is only 0.1, so the statistically significant difference is not large enough that we really care. Statistical significance doesn't mean it's also an important difference. The TOST test strongly rejects that there is a difference of at least 0.5, indicating that given our threshold of 0.5, the two strategies are the same.
import numpy as np
from scipy import stats
import statsmodels.stats.weightstats as smws
nobs_all = [10, 100, 1000]
sigma = 0.5
seed = 628561 #chosen to produce nice result in small sample
print seed
for nobs in nobs_all:
np.random.seed(seed)
payoff_s1 = sigma * np.random.randn(nobs)
payoff_s2 = 0.1 + sigma * np.random.randn(nobs)
p1 = stats.ttest_ind(payoff_s1, payoff_s2)[1]
p2 = smws.tost_ind(payoff_s1, payoff_s2, -0.5, 0.5, usevar='pooled')[0]
print 'nobs:', nobs, 'diff in means:', payoff_s2.mean() - payoff_s1.mean()
print 'ttest:', p1, ['not different', 'different '][p1 < 0.05],
print ' tost:', p2, ['different ', 'not different'][p2 < 0.05]
But from your description of QR, I thought specifically of the case
where we have a "natural" ordering of the regressors, similar to the
polynomial case of you and Anne. In the timeseries case, it would be by
increasing lags
yt on y_{t-1}
yt on y_{t-1}, y_{t-2}
...
...
yt on y_{t-k} for k= 1,...,K
or yt on xt and the lags of xt
This is really sequential LS with a predefined sequence, not PLS or
PCA/PCR or similar orthogonalization by "importance".
The usual procedure for deciding on the appropriate number of lags
usually loops over OLS with increasing number of regressors.
>From the discussion, I thought there might be a way to "cheat" in this
using QR and Gram-Schmidt
I never got around trying this out, but I thought I give it a try today.
Some hours of trial and error and working on some algebra later, I have
what looks like a reasonable solution.import numpy as np from statsmodels.regression.linear_model import OLS from numpy.testing import assert_almost_equalThen, we define a toy example to try out whether it works. The last two variables have no effect. I used that to get the minimum of the information criteria, aic, bic, to be in interior.
nobs, k_vars = 50, 4 np.random.seed(85325783) x = np.random.randn(nobs, k_vars) y = x[:, :k_vars-2].sum(1) + np.random.randn(nobs)We start with the boring way of doing the sequential regression: use OLS from statsmodels and loop with expanding number of explanatory variables in exog.
ssr_ols = np.zeros(k_vars)
params_ols = np.zeros((k_vars, k_vars))
ic_ols = np.zeros((2, k_vars))
for i in range(4):
res = OLS(y, x[:,:i+1]).fit()
ssr_ols[i] = res.ssr
params_ols[i, :i+1] = res.params
ic_ols[0, i] = res.aic
ic_ols[1, i] = res.bic
In my example, the estimated coefficients are>>> params_ols
array([[ 0.7585129 , 0. , 0. , 0. ],
[ 1.00564191, 0.96414302, 0. , 0. ],
[ 1.00776594, 0.93035613, -0.10759121, 0. ],
[ 1.03379054, 0.91302697, -0.12998046, -0.08787965]])
The first two coefficients are close to one, the second two coefficients are close to zero, all in the neighborhood of the true values of my simulated model.q, r = np.linalg.qr(x) qy = np.dot(q.T, y) print '\nparams full model' print params_ols[-1] print np.linalg.solve(r, qy)It gives the same result as the pinv version
params full model [ 1.03379054 0.91302697 -0.12998046 -0.08787965] [ 1.03379054 0.91302697 -0.12998046 -0.08787965]We already have the QR decomposition for the full model. QR` calculates the orthogonalization using the sequence as it is given by the matrix of explanatory variables.
params1 = np.zeros((k_vars, k_vars))
for i in range(4):
params1[i, :i+1] = np.linalg.solve(r[:i+1, :i+1], qy[:i+1])
This gives us the same parameter estimates as the OLS loop. The computations
are on much smaller arrays than the original problem, r is (k_vars, k_vars)
and qy is just one dimensional with length k_vars.All we need to do is to select the sequentially increasing subsets of r and theta. For calculating predicted values, we can work directly in the orthogonalized system.
- q is the orthogonal matrix that spans the same space as x.
- theta = r beta = qy is the parameter vector in the orthogonalized problem.
- beta = inv(r) theta is the parameter vector for the original regressors.
upp_tri = (np.arange(4)[:,None] <= np.arange(4)).astype(float)we can get the expanding theta or qy in a matrix
>>> upp_tri * qy[:,None]
array([[ 4.87045847, 4.87045847, 4.87045847, 4.87045847],
[-0. , -6.94085588, -6.94085588, -6.94085588],
[ 0. , 0. , 0.70410517, 0.70410517],
[ 0. , 0. , 0. , 0.60930516]])
Then, we can just solve the system for all steps at once. First, I tried
the matrix inverse since that's easier to understand theoreticallyparams2 = np.dot(np.linalg.inv(r), upp_tri * qy[:,None]).TBut, we can avoid the inverse and use linalg.solve directly:
params2 = np.linalg.solve(r, upp_tri * qy[:,None]).TThat's it, one line and I have the parameters for the four sequential regression problems. Comparing the parameter estimates of the three methods, we see they are the same (at floating point precision)
>>> np.max(np.abs(params1 - params2)) 0.0 >>> np.max(np.abs(params1 - params_ols)) 5.5511151231257827e-16The rest is calculating fitted values and residuals
contrib = q * qy fitted = contrib.cumsum(1) resids = y[:,None] - fitted ssrs = (resids**2).sum(0) print '\nresidual sum of squares ssr' print ssrs print ssr_olswhich gives us identical residual sum of squares, ssr
[ 80.31708213 32.14160173 31.64583764 31.27458486] [ 80.31708213 32.14160173 31.64583764 31.27458486]I added some asserts in the script, so I don't break anything by accident
assert_almost_equal(ssrs, ssr_ols, decimal=13) assert_almost_equal(params1, params_ols, decimal=13) assert_almost_equal(params2, params_ols, decimal=13)The applications, that I have in mind for this, are selection of the number of regressors or lags in the time series case, so I want to calculate the information criteria, aic and bic. In the previous result, I had calculated the residual sum of squares, which I can feed to helper functions that I had written to calculate information criteria.
print '\naic, bic' print ic_ols import statsmodels.tools.eval_measures as evm dfmodelwc = range(1, k_vars+1) #number of variables in regression aics = [evm.aic_sigma(ssrs[i], nobs, dfmodelwc[i]) for i in range(k_vars)] bics = [evm.bic_sigma(ssrs[i], nobs, dfmodelwc[i]) for i in range(k_vars)] print aics print bics print 'aic, bic minimum number of variables' print np.argmin(ic_ols, 1) + 1 print np.argmin(aics) + 1, np.argmin(aics) + 1prints
aic, bic [[ 167.59181941 123.80026281 125.02303444 126.43299217] [ 169.50384241 127.62430882 130.75910345 134.08108419]] [4.4259823271765022, 3.5501511951835187, 3.5746066277468964, 3.6028057824070068] [4.4642227872850651, 3.6266321154006445, 3.689328008072585, 3.7557676228412582] aic, bic minimum number of variables [2 2] 2 2PS: Since I never had any numerical linear algebra training, I can still get excited about figuring out a two-liner. I could have tried to find a book for it, but that would have been boring compared to working it out on my own.
>>> print res.summary()
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.901
Model: OLS Adj. R-squared: 0.898
Method: Least Squares F-statistic: 290.3
Date: Thu, 10 May 2012 Prob (F-statistic): 5.31e-48
Time: 13:15:22 Log-Likelihood: -173.85
No. Observations: 100 AIC: 355.7
Df Residuals: 96 BIC: 366.1
Df Model: 3
==============================================================================
coef std err t P>|t| [95.0% Conf. Int.]
------------------------------------------------------------------------------
x1 0.4872 0.024 20.076 0.000 0.439 0.535
x2 0.5408 0.045 12.067 0.000 0.452 0.630
x3 0.5136 0.030 16.943 0.000 0.453 0.574
const 4.6294 0.372 12.446 0.000 3.891 5.368
==============================================================================
Omnibus: 0.945 Durbin-Watson: 1.570
Prob(Omnibus): 0.624 Jarque-Bera (JB): 1.031
Skew: -0.159 Prob(JB): 0.597
Kurtosis: 2.617 Cond. No. 33.2
==============================================================================
The following three graphs are refactored versions of the regression plots. Each graph looks at the data and estimation results with respect to one of the three variables. (The graphs look better in original size.)
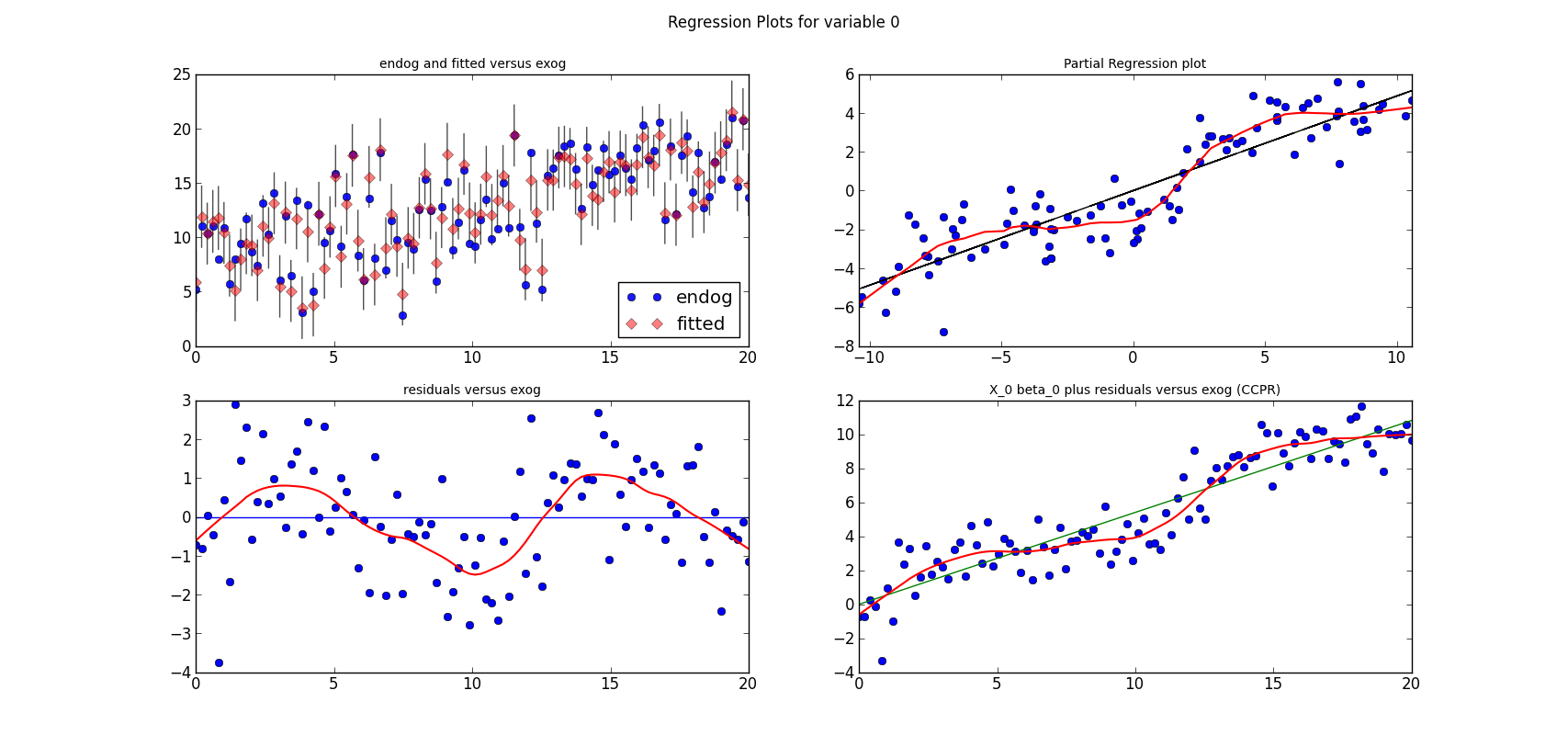
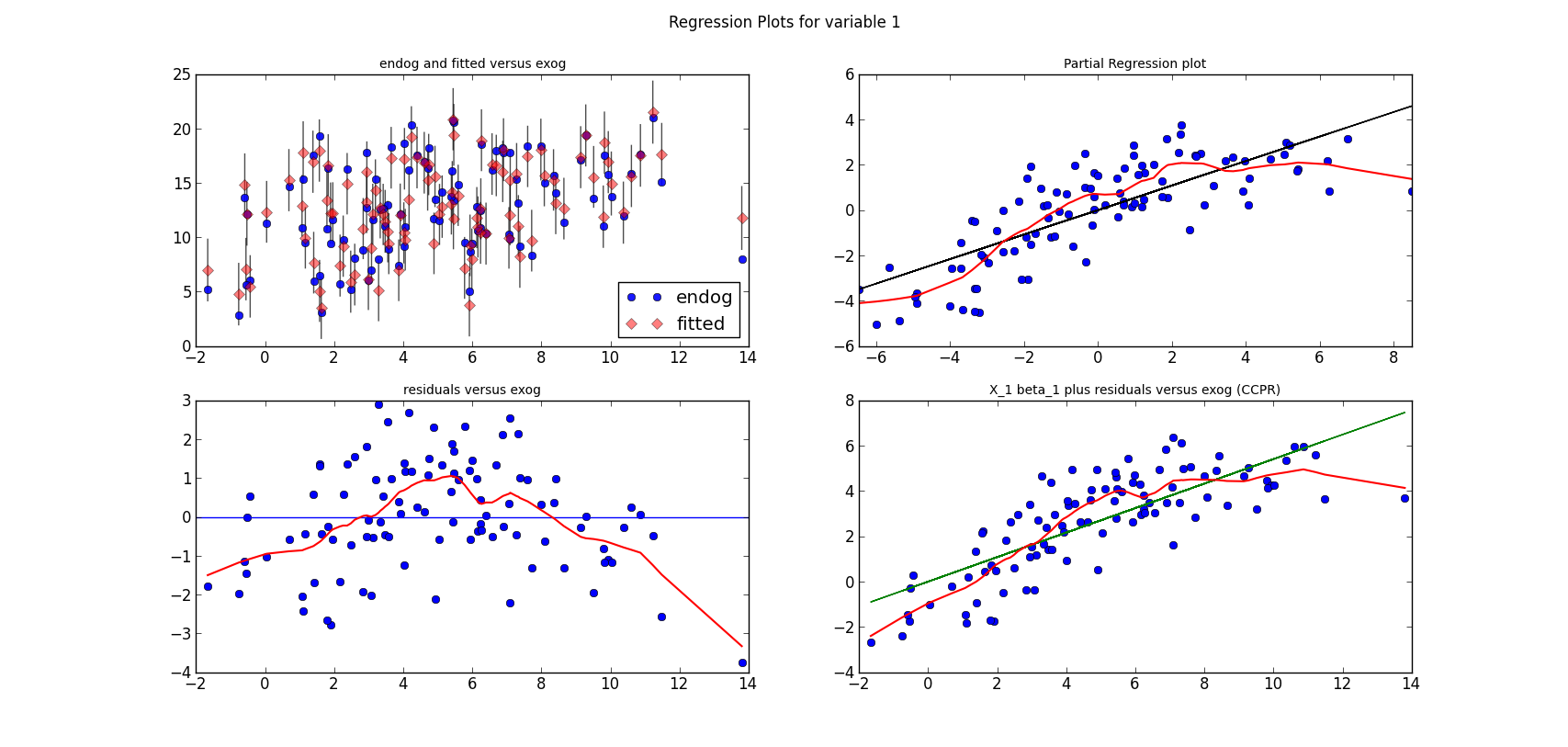
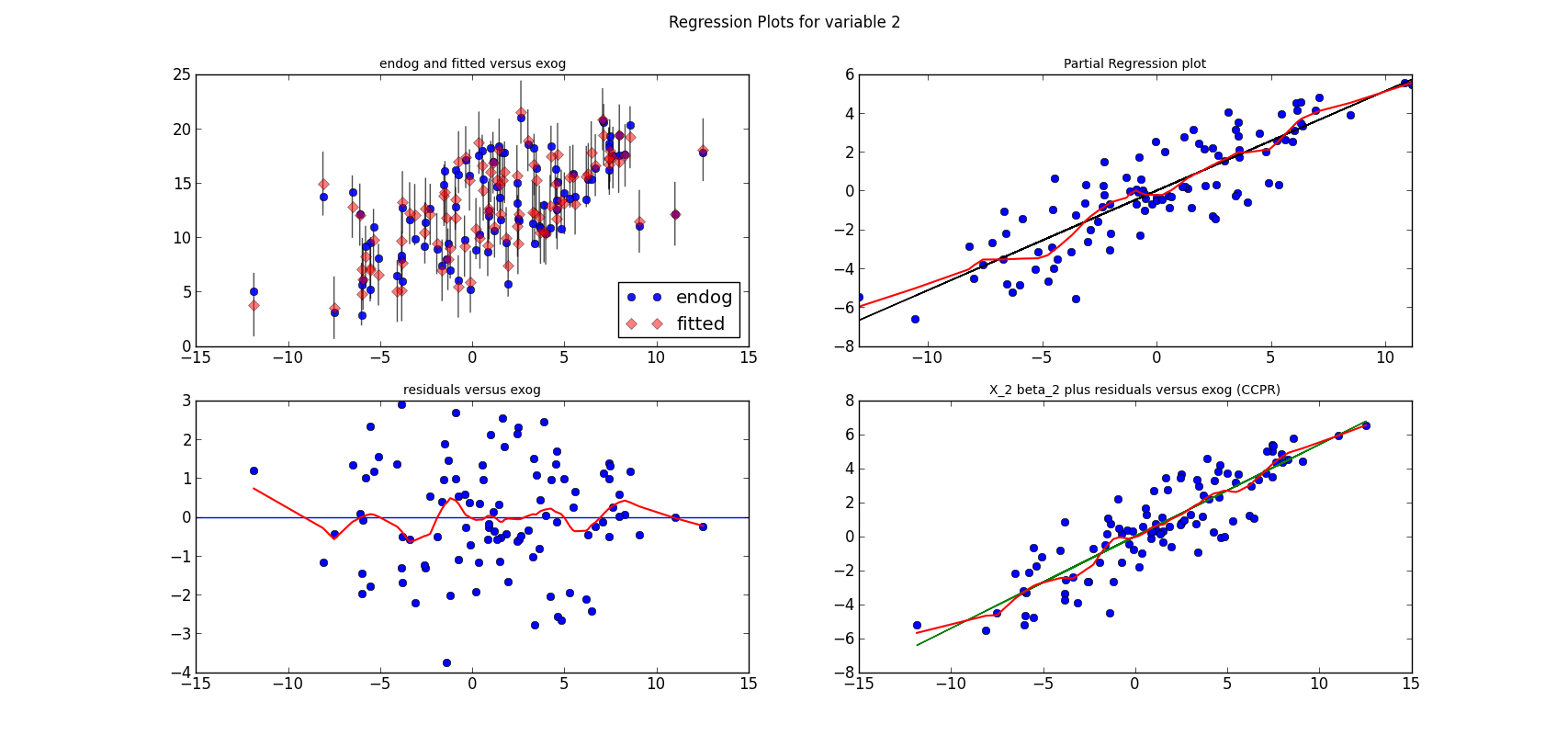
from regressionplots_new import plot_regress_exog fig9 = plot_regress_exog(res, exog_idx=0) add_lowess(fig9, ax_idx=1, lines_idx=0) add_lowess(fig9, ax_idx=2, lines_idx=0) add_lowess(fig9, ax_idx=3, lines_idx=0) fig10 = plot_regress_exog(res, exog_idx=1) add_lowess(fig10, ax_idx=1, lines_idx=0) add_lowess(fig10, ax_idx=2, lines_idx=0) add_lowess(fig10, ax_idx=3, lines_idx=0) fig11 = plot_regress_exog(res, exog_idx=2) add_lowess(fig11, ax_idx=1, lines_idx=0) add_lowess(fig11, ax_idx=2, lines_idx=0) add_lowess(fig11, ax_idx=3, lines_idx=0)
statsmodels has a graphics subdirectory, where we started to collect some of the common statistical plots. To make the documentation a bit more exciting, I am adding plots directly to the docstrings for the individual functions. Currently, we don't have many of them in the online documentation yet, two examples violin_plot and bean_plot.
qqplot(data, dist=stats.norm, distargs=(), a=0, loc=0, scale=1, fit=False, line=False, ax=None)I am not copying the entire docstring, what I would like to present here are some examples and how to work with the plots.
's' - standardized line, the expected order statistics are scaled by the standard deviation of the given sample and have the mean added to themcorresponds to the line after fitting location and scale for the normal distribution
- The first plot fits a normal distribution, keywords: line='45', fit=True
- The second plot fits the t distribution, keywords: dist=stats.t, line='45', fit=True
- The third plot is the same as the second plot, but I fit the t distribution myself, instead of having qqplot do it. keywords: dist=stats.t, distargs=(dof,), loc=loc, scale=scale, line='45'. I added the estimated parameters into a text insert in the plot. qqplot currently doesn't tell us what the fitted parameters are.
from scipy import stats import statsmodels.api as sm #estimate to get the residuals data = sm.datasets.longley.load() data.exog = sm.add_constant(data.exog) mod_fit = sm.OLS(data.endog, data.exog).fit() res = mod_fit.resid fig = sm.graphics.qqplot(res, dist=stats.t, line='45', fit=True) fig.show()It works but the x-axis goes from -3 to 3, while there are only values from -2 to 2.
fig.axes[0].set_xlim(-2, 2)The resulting plot is then the same as the third plot in the first graph above.
from scipy import stats from matplotlib import pyplot as plt import statsmodels.api as sm #example from docstring data = sm.datasets.longley.load() data.exog = sm.add_constant(data.exog) mod_fit = sm.OLS(data.endog, data.exog).fit() res = mod_fit.residThen I hardcode a left position for text inserts, and create a matplotlib figure instance
left = -1.8 fig = plt.figure()Next we can add the first subplot. The only keyword arguments for qqplot is ax to tell qqplot to attach the plot to my first subplot. Since I want to insert a text to describe the keywords, I needed to spend some time with the matplotlib documentation. As we have a reference to the axis instance, it is easy to change or add plot elements
ax = fig.add_subplot(2, 2, 1) sm.graphics.qqplot(res, ax=ax) top = ax.get_ylim()[1] * 0.75 txt = ax.text(left, top, 'no keywords', verticalalignment='top') txt.set_bbox(dict(facecolor='k', alpha=0.1))The other subplots follow the same pattern. I didn't try to generalize or avoid hardcoding
ax = fig.add_subplot(2, 2, 2)
sm.graphics.qqplot(res, line='s', ax=ax)
top = ax.get_ylim()[1] * 0.75
txt = ax.text(left, top, "line='s'", verticalalignment='top')
txt.set_bbox(dict(facecolor='k', alpha=0.1))
ax = fig.add_subplot(2, 2, 3)
sm.graphics.qqplot(res, line='45', fit=True, ax=ax)
ax.set_xlim(-2, 2)
top = ax.get_ylim()[1] * 0.75
txt = ax.text(left, top, "line='45', \nfit=True", verticalalignment='top')
txt.set_bbox(dict(facecolor='k', alpha=0.1))
ax = fig.add_subplot(2, 2, 4)
sm.graphics.qqplot(res, dist=stats.t, line='45', fit=True, ax=ax)
ax.set_xlim(-2, 2)
top = ax.get_ylim()[1] * 0.75
txt = ax.text(left, top, "dist=stats.t, \nline='45', \nfit=True",
verticalalignment='top')
txt.set_bbox(dict(facecolor='k', alpha=0.1))
The final step is to adjust the layout, so that axis labels don't overlap with other subplots if the graph is not very largefig.tight_layout()
import numpy as np seed = np.random.randint(1000000) print 'seed', seed seed = 461970 #nice seed for nobs=1000 #seed = 571478 #nice seed for nobs=100 #seed = 247819 #for nobs=100, estimated t is essentially normal np.random.seed(seed) rvs = stats.t.rvs(4, size=1000)The first two subplot are very similar to what is in the first graph
fig2 = plt.figure() ax = fig2.add_subplot(2, 2, 1) fig2 = sm.graphics.qqplot(rvs, dist=stats.norm, line='45', fit=True, ax=ax) top = ax.get_ylim()[1] * 0.75 xlim = ax.get_xlim() frac = 0.1 left = xlim[0] * (1-frac) + xlim[1] * frac txt = ax.text(left, top, "normal", verticalalignment='top') txt.set_bbox(dict(facecolor='k', alpha=0.1)) ax = fig2.add_subplot(2, 2, 2) fig2 = sm.graphics.qqplot(rvs, dist=stats.t, line='45', fit=True, ax=ax) top = ax.get_ylim()[1] * 0.75 xlim = ax.get_xlim() frac = 0.1 left = xlim[0] * (1-frac) + xlim[1] * frac txt = ax.text(left, top, "t", verticalalignment='top') txt.set_bbox(dict(facecolor='k', alpha=0.1))For the third plot, I estimate the parameters of the t-distribution to see whether I get the same results as in the second plot (I do), and so I can insert the parameter estimates into the plot
params = stats.t.fit(rvs)
dof, loc, scale = params
ax = fig2.add_subplot(2, 2, 4)
fig2 = sm.graphics.qqplot(rvs, dist=stats.t, distargs=(dof,), loc=loc,
scale=scale, line='45', fit=False, ax=ax)
top = ax.get_ylim()[1] * 0.75
xlim = ax.get_xlim()
frac = 0.1
left = xlim[0] * (1-frac) + xlim[1] * frac
txt = ax.text(left, top, "t \ndof=%3.2F \nloc=%3.2F, \nscale=%3.2F" % tuple(params),
verticalalignment='top')
txt.set_bbox(dict(facecolor='k', alpha=0.1))
That's it for the plots, now I need to add them to the statsmodels documentation.>>> normal_ad(res) (0.43982328207860633, 0.25498161947268855) >>> lillifors(res) (0.17229856392873188, 0.2354638181341876)On the other hand, in the second example with 1000 observations from the t distribution, the assumption that the data comes from a normal distribution is clearly rejected
>>> normal_ad(rvs) (6.5408483355136013, 4.7694160497092537e-16) >>> lillifors(rvs) (0.05919821253474411, 8.5872265678140885e-09)PPS:
>>> from statsmodels.stats.adnorm import normal_ad >>> from statsmodels.stats.lilliefors import lillifors
I didn't have much time or motivation to work on my blog these last weeks, mainly because I was busy discussing Google Summer of Code and preparing a new release for statsmodels.
So here is just an update on our Google Summer of Code candidates and their projects. This year was a successful year in attracting student proposals. We have six proposals, five of them we discussed quite extensively on our mailing list before the application.
Of the five projects, the first two are must-haves for econometrics or statistical packages, one on System of Equations, the other on Nonlinear Least-Squares and Nonlinear Robust Models. The next two are nonparametric or semi-parametric methods, one more traditional kernel estimation, the other using Empirical Likelihood which is a relatively new approach that has become popular in recent research both in econometrics and in statistics. The fifth is on Dynamic Linear Models mainly using Kalman filter and a Bayesian approach, which would extend the depth of statsmodels in time series analysis.
All topics would be valuable extensions to statsmodels and significantly increase our coverage of statistics and econometrics. From the discussion on the mailing list I think that all candidates are qualified to bring the projects to a successful finish.
This is a standard econometrics topic, but I only recently found that graphical models and causal models discussed in other fields have a large overlap with this. In the case of a system of simultaneous equations, we have several variables that depend on each other. The simplest case in economics is a market equilibrium, where the demanded and supplied quantities depend on the price, and the price depends on the supply and demand. The estimation methods commonly used in this area are two-stage and three-stage least-squares and limited and full information maximum likelihood estimation. The first part of the project starts with the simpler case when we have several response variables, but they don't depend on each other simultaneously, although they can depend on past values of other response variables. I'm very glad that someone is picking this one up.
This project has two parts, the first is extending the linear least-squares model to the non-linear case, the second part is to implement non-linear models for robust estimation. Non-linear least squares is available in scipy for example with scipy.optimize.curve_fit. However, in the statsmodels version we want to provide all the usual results statistics and statistical tests. The second part will implement two robust estimators for non-linear model, that have been shown to be most successful in recent Monte Carlo studies comparing different robust estimators for non-linear equations. Robust estimation here refers to the case when there are possibly many outliers in the data. My guess is that these will become the most used models of all proposals.
This project extends the kernel based method in statsmodels from the univariate to the multivariate case, will provide better bandwidth selection, and then implement nonparametric function estimation. Multivariate kernel density estimation should complement scipy.stats.gaussian_kde which only works well with distributions that are approximately normal shaped or have only a single peak. Another extension is to provide kernels and estimation methods for discrete variables. These methods have been on our wishlist for a while, but only the univariate case has been included in statsmodels so far.
This is a relatively new approach in statistics and econometrics that avoids the distributional assumptions in estimation and in statistical tests. Instead of relying on a known distribution in small samples, where we often assume normal distribution, or instead of relying on the asymptotic normal distribution in large samples, this approach estimates the distribution in a nonparametric way. This is similar, to some extend, to the difference between, for example, a t-test and a rank-based Mann–Whitney U or Wilcoxon test, which are available in scipy.stats. The advantages are that in smaller samples the estimates and tests are more accurate when the distribution is not known, and in many cases, for example in finance, most tests show that the assumption of normal distribution does not hold. For this project, I still have to catch up with some readings because I'm only familiar with a small part of this literature, mainly on empirical likelihood in relation to Generalized Method of Moments (GMM).
This covers statespace models implemented by Kalman Filter for multivariate time series models, both from a likelihood and a Bayesian perspective. The project expands the coverage of statsmodels in linear time series analysis, the first area where we get a good coverage of models. Currently, we have univariate AR and ARIMA, vector autoregressive models VAR, and structural VAR. Part of this project would be to get a good cython based implementation of Kalman filters. Wes has started a libray, statlib, for this last year, however, it is still incomplete and needs to be integrated with statsmodels. Another advantage of this project is that it increases our infrastructure and models for macro-econometrics, estimation of macroeconomic models and dynamic stochastic general equilibrium DSGE models, which is currently still Matlab dominated, as far as I can tell.
Now we still have to see how many GSoC slots we will get, but we have the chance this year to get a large increase in the speed of development of statsmodels, and we can reduce the number of cases where someone needs to run to R, or Stata, or Matlab because there is no implementation for a statistical analysis available in Python.