I just want to show another two plots for the statistical power of a test, since I didn't have time for this earlier
The code to produce them is just calling the methods of the power classes, for example for the one sample t-test.
I just want to show another two plots for the statistical power of a test, since I didn't have time for this earlier
The code to produce them is just calling the methods of the power classes, for example for the one sample t-test.
>>> print res.summary()
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.901
Model: OLS Adj. R-squared: 0.898
Method: Least Squares F-statistic: 290.3
Date: Thu, 10 May 2012 Prob (F-statistic): 5.31e-48
Time: 13:15:22 Log-Likelihood: -173.85
No. Observations: 100 AIC: 355.7
Df Residuals: 96 BIC: 366.1
Df Model: 3
==============================================================================
coef std err t P>|t| [95.0% Conf. Int.]
------------------------------------------------------------------------------
x1 0.4872 0.024 20.076 0.000 0.439 0.535
x2 0.5408 0.045 12.067 0.000 0.452 0.630
x3 0.5136 0.030 16.943 0.000 0.453 0.574
const 4.6294 0.372 12.446 0.000 3.891 5.368
==============================================================================
Omnibus: 0.945 Durbin-Watson: 1.570
Prob(Omnibus): 0.624 Jarque-Bera (JB): 1.031
Skew: -0.159 Prob(JB): 0.597
Kurtosis: 2.617 Cond. No. 33.2
==============================================================================
The following three graphs are refactored versions of the regression plots. Each graph looks at the data and estimation results with respect to one of the three variables. (The graphs look better in original size.)
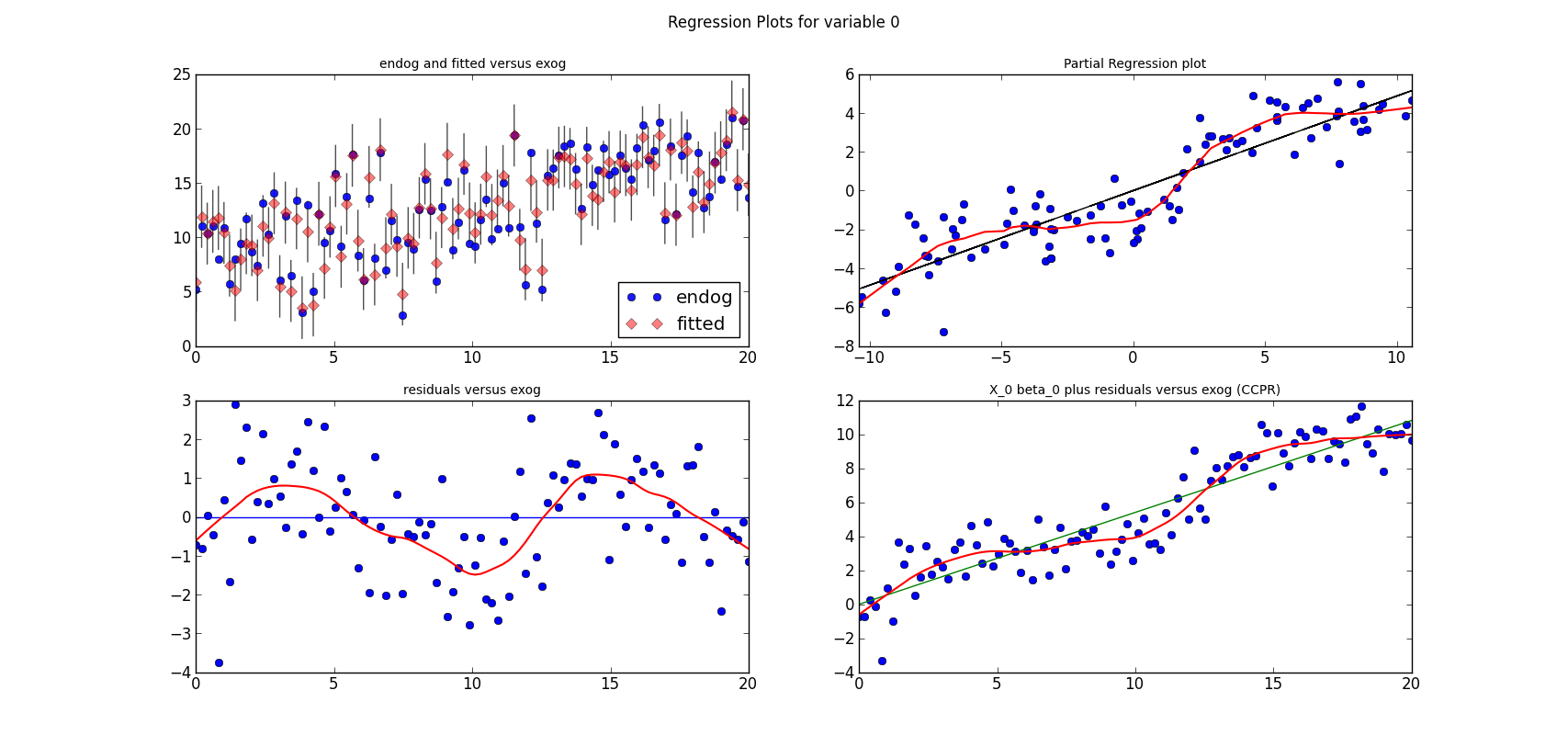
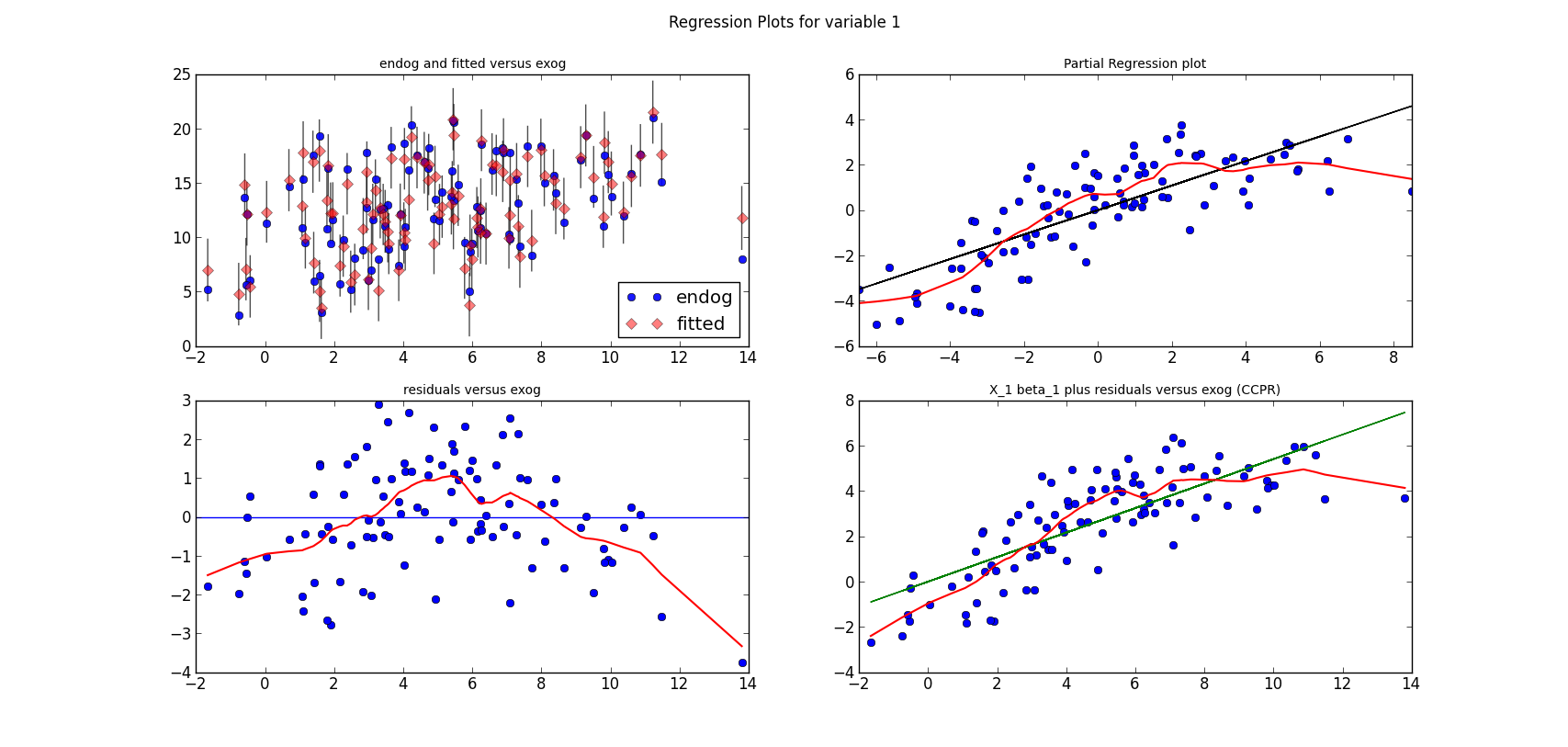
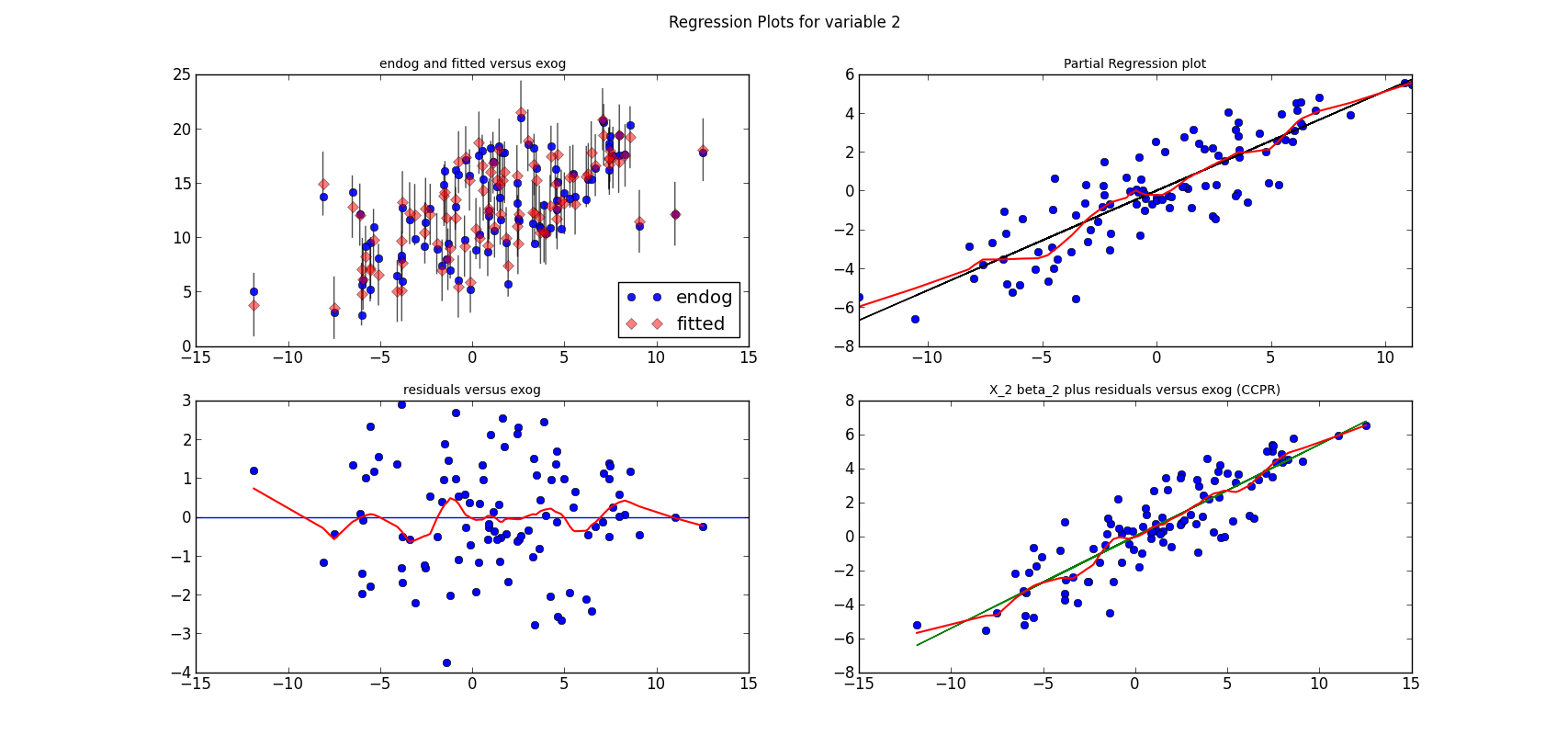
from regressionplots_new import plot_regress_exog fig9 = plot_regress_exog(res, exog_idx=0) add_lowess(fig9, ax_idx=1, lines_idx=0) add_lowess(fig9, ax_idx=2, lines_idx=0) add_lowess(fig9, ax_idx=3, lines_idx=0) fig10 = plot_regress_exog(res, exog_idx=1) add_lowess(fig10, ax_idx=1, lines_idx=0) add_lowess(fig10, ax_idx=2, lines_idx=0) add_lowess(fig10, ax_idx=3, lines_idx=0) fig11 = plot_regress_exog(res, exog_idx=2) add_lowess(fig11, ax_idx=1, lines_idx=0) add_lowess(fig11, ax_idx=2, lines_idx=0) add_lowess(fig11, ax_idx=3, lines_idx=0)
statsmodels has a graphics subdirectory, where we started to collect some of the common statistical plots. To make the documentation a bit more exciting, I am adding plots directly to the docstrings for the individual functions. Currently, we don't have many of them in the online documentation yet, two examples violin_plot and bean_plot.
qqplot(data, dist=stats.norm, distargs=(), a=0, loc=0, scale=1, fit=False, line=False, ax=None)I am not copying the entire docstring, what I would like to present here are some examples and how to work with the plots.
's' - standardized line, the expected order statistics are scaled by the standard deviation of the given sample and have the mean added to themcorresponds to the line after fitting location and scale for the normal distribution
- The first plot fits a normal distribution, keywords: line='45', fit=True
- The second plot fits the t distribution, keywords: dist=stats.t, line='45', fit=True
- The third plot is the same as the second plot, but I fit the t distribution myself, instead of having qqplot do it. keywords: dist=stats.t, distargs=(dof,), loc=loc, scale=scale, line='45'. I added the estimated parameters into a text insert in the plot. qqplot currently doesn't tell us what the fitted parameters are.
from scipy import stats import statsmodels.api as sm #estimate to get the residuals data = sm.datasets.longley.load() data.exog = sm.add_constant(data.exog) mod_fit = sm.OLS(data.endog, data.exog).fit() res = mod_fit.resid fig = sm.graphics.qqplot(res, dist=stats.t, line='45', fit=True) fig.show()It works but the x-axis goes from -3 to 3, while there are only values from -2 to 2.
fig.axes[0].set_xlim(-2, 2)The resulting plot is then the same as the third plot in the first graph above.
from scipy import stats from matplotlib import pyplot as plt import statsmodels.api as sm #example from docstring data = sm.datasets.longley.load() data.exog = sm.add_constant(data.exog) mod_fit = sm.OLS(data.endog, data.exog).fit() res = mod_fit.residThen I hardcode a left position for text inserts, and create a matplotlib figure instance
left = -1.8 fig = plt.figure()Next we can add the first subplot. The only keyword arguments for qqplot is ax to tell qqplot to attach the plot to my first subplot. Since I want to insert a text to describe the keywords, I needed to spend some time with the matplotlib documentation. As we have a reference to the axis instance, it is easy to change or add plot elements
ax = fig.add_subplot(2, 2, 1) sm.graphics.qqplot(res, ax=ax) top = ax.get_ylim()[1] * 0.75 txt = ax.text(left, top, 'no keywords', verticalalignment='top') txt.set_bbox(dict(facecolor='k', alpha=0.1))The other subplots follow the same pattern. I didn't try to generalize or avoid hardcoding
ax = fig.add_subplot(2, 2, 2)
sm.graphics.qqplot(res, line='s', ax=ax)
top = ax.get_ylim()[1] * 0.75
txt = ax.text(left, top, "line='s'", verticalalignment='top')
txt.set_bbox(dict(facecolor='k', alpha=0.1))
ax = fig.add_subplot(2, 2, 3)
sm.graphics.qqplot(res, line='45', fit=True, ax=ax)
ax.set_xlim(-2, 2)
top = ax.get_ylim()[1] * 0.75
txt = ax.text(left, top, "line='45', \nfit=True", verticalalignment='top')
txt.set_bbox(dict(facecolor='k', alpha=0.1))
ax = fig.add_subplot(2, 2, 4)
sm.graphics.qqplot(res, dist=stats.t, line='45', fit=True, ax=ax)
ax.set_xlim(-2, 2)
top = ax.get_ylim()[1] * 0.75
txt = ax.text(left, top, "dist=stats.t, \nline='45', \nfit=True",
verticalalignment='top')
txt.set_bbox(dict(facecolor='k', alpha=0.1))
The final step is to adjust the layout, so that axis labels don't overlap with other subplots if the graph is not very largefig.tight_layout()
import numpy as np seed = np.random.randint(1000000) print 'seed', seed seed = 461970 #nice seed for nobs=1000 #seed = 571478 #nice seed for nobs=100 #seed = 247819 #for nobs=100, estimated t is essentially normal np.random.seed(seed) rvs = stats.t.rvs(4, size=1000)The first two subplot are very similar to what is in the first graph
fig2 = plt.figure() ax = fig2.add_subplot(2, 2, 1) fig2 = sm.graphics.qqplot(rvs, dist=stats.norm, line='45', fit=True, ax=ax) top = ax.get_ylim()[1] * 0.75 xlim = ax.get_xlim() frac = 0.1 left = xlim[0] * (1-frac) + xlim[1] * frac txt = ax.text(left, top, "normal", verticalalignment='top') txt.set_bbox(dict(facecolor='k', alpha=0.1)) ax = fig2.add_subplot(2, 2, 2) fig2 = sm.graphics.qqplot(rvs, dist=stats.t, line='45', fit=True, ax=ax) top = ax.get_ylim()[1] * 0.75 xlim = ax.get_xlim() frac = 0.1 left = xlim[0] * (1-frac) + xlim[1] * frac txt = ax.text(left, top, "t", verticalalignment='top') txt.set_bbox(dict(facecolor='k', alpha=0.1))For the third plot, I estimate the parameters of the t-distribution to see whether I get the same results as in the second plot (I do), and so I can insert the parameter estimates into the plot
params = stats.t.fit(rvs)
dof, loc, scale = params
ax = fig2.add_subplot(2, 2, 4)
fig2 = sm.graphics.qqplot(rvs, dist=stats.t, distargs=(dof,), loc=loc,
scale=scale, line='45', fit=False, ax=ax)
top = ax.get_ylim()[1] * 0.75
xlim = ax.get_xlim()
frac = 0.1
left = xlim[0] * (1-frac) + xlim[1] * frac
txt = ax.text(left, top, "t \ndof=%3.2F \nloc=%3.2F, \nscale=%3.2F" % tuple(params),
verticalalignment='top')
txt.set_bbox(dict(facecolor='k', alpha=0.1))
That's it for the plots, now I need to add them to the statsmodels documentation.>>> normal_ad(res) (0.43982328207860633, 0.25498161947268855) >>> lillifors(res) (0.17229856392873188, 0.2354638181341876)On the other hand, in the second example with 1000 observations from the t distribution, the assumption that the data comes from a normal distribution is clearly rejected
>>> normal_ad(rvs) (6.5408483355136013, 4.7694160497092537e-16) >>> lillifors(rvs) (0.05919821253474411, 8.5872265678140885e-09)PPS:
>>> from statsmodels.stats.adnorm import normal_ad >>> from statsmodels.stats.lilliefors import lillifors